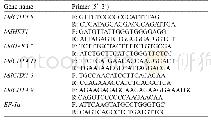
《Table 1 Genes and primers used in the quantitative real-time PCR analysis》
 提示:宽带有限、当前游客访问压缩模式
提示:宽带有限、当前游客访问压缩模式
本系列图表出处文件名:随高清版一同展现
《Primary molecular basis of androgenic gland endocrine sex regulation revealed by transcriptome analysis in Eriocheir sinensis》
The q RT-PCR was performed used new AG_ED and ED samples to verify the accuracy of RNA-Seq and the expression level of key DEGs.Total RNA was extracted and the fi rst-strand cDNA was synthesized by using Prime Script RT Reagent Kit with g DNA Eraser(TaKaRa,Japan).Then,the cDNA was diluted100 times by nucleic acid free water.SYBR Green Real-time RT-PCR was performed in an ABI 7500Sequence Detection System(Applied Biosystems).Nine pairs of gene-specifi c primers(Table 1)were used to amplify the partial c DNA sequences,respectively.Theβ-actin gene was amplifi ed in parallel as an internal control.There were three biological and three technical replicates respectively.The 2-ΔΔC\n\t\t\t\t\tt method was used to analyze the expression level of diff erent genes(Livak and Schmittgen,2001).Simply,the fold change in target gene relative to theβ-actin endogenous control is determined by:Fold change=2-ΔΔC\n\t\t\t\t\tt,where the C tvalue is defi ned as the cycle number at which the fl uorescence signal crosses the threshold;
| 图表编号 | XD0034432100 严禁用于非法目的 |
|---|---|
| 绘制时间 | 2019.01.01 |
| 作者 | SONG Chengwen、LIU Lei、HUI Min、LIU Yuan、LIU Hourong、CUI Zhaoxia |
| 绘制单位 | CAS Key Laboratory of Experimental Marine Biology, Institute of Oceanology, Chinese Academy of Sciences、Laboratory for Marine Biology and Biotechnology, Qingdao National Laboratory for Marine Science and Technology、CAS Key Laboratory of Experimental Marin |
| 更多格式 | 高清、无水印(增值服务) |