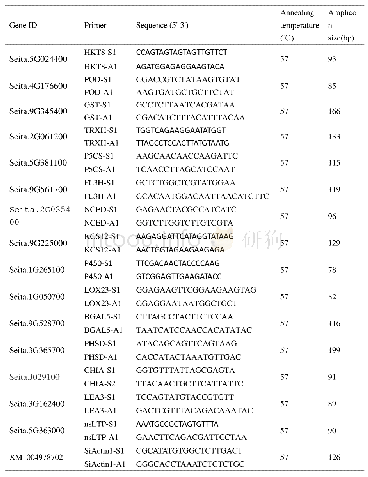
《Tab.1 Sequences of the primers for amplifying the genes using real-time RT-PCR》
 提示:宽带有限、当前游客访问压缩模式
提示:宽带有限、当前游客访问压缩模式
本系列图表出处文件名:随高清版一同展现
《地塞米松对内毒素诱导的小鼠葡萄膜炎的治疗效果及转录组分析(英文)》
The total RNA of neural retina was extracted with Trizol Reagent(Invitrogen,Carlsbad,CA,USA)following the manufacturer's instructions.For quantitative PCR,c DNA was synthesized using the Prime Script RT reagent kit(TakaraBiotechnology,Dalian,China).PCR amplification was performed using a real-time PCR system(ABI 7500 system,Applied Biosystems,Foster City,CA,USA)with SYBR Green(Vazyme Biotech Co.,Ltd,Nanjing,China).The thermal cycles were carried out with initial denaturation at 95℃for 10 min,followed by40 cycles of 95℃for 15 s and 60℃for 1 mi.To determine the m RNA levels,GAPDH was used as the endogenous reference gene.All the samples were prepared in duplicate and the average Ct values were used for quantification.Relative quantification was achieved using the comparative 2-ΔΔCtmethod.The control group was used as the calibrator to calculate the relative fold change of the m RNA of the target gene X in the experimental group(EG):ΔΔCt=(Ct.XEG-Ct.GAPDHEG)/(Ct.Xcontrol-Ct.GAPDHcontrol)[21].The sequences of primers and Gen Bank accession numbers of the target genes are listed in Tab.1.
| 图表编号 | XD0016638600 严禁用于非法目的 |
|---|---|
| 绘制时间 | 2018.08.20 |
| 作者 | 余鹏、邱一果、林茹、符馨予、郝冰涛、雷博 |
| 绘制单位 | 重庆医科大学附属第一医院眼科、、眼科学重庆市市级重点实验室、、重庆市眼科研究所、重庆医科大学附属第一医院眼科、、眼科学重庆市市级重点实验室、、重庆市眼科研究所、重庆医科大学附属第一医院眼科、、眼科学重庆市市级重点实验室、、重庆市眼科研究所、重庆医科大学附属第一医院眼科、、眼科学重庆市市级重点实验室、、重庆市眼科研究所、南方医科大学癌症研究所、重庆医科大学附属第一医院眼科、、眼科学重庆市市级重点实验室、、重庆市眼科研究所、河南省人民医院河南省立眼科研究所 |
| 更多格式 | 高清、无水印(增值服务) |