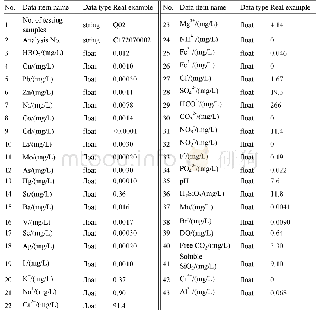
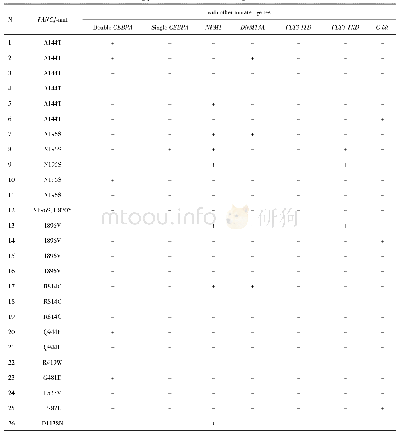
《Table 2.Mutations and segregation analysis of the TKC1.2-LAZY1 T1 plants.》
 提示:宽带有限、当前游客访问压缩模式
提示:宽带有限、当前游客访问压缩模式
本系列图表出处文件名:随高清版一同展现
《Improvements of TKC Technology Accelerate Isolation of Transgene-Free CRISPR/Cas9-Edited Rice Plants》
Protospacer adjacent motif site AGG is underlined.Zhonghua 11 plants were used as wild-type(WT).‘+’refers to base pair insertion.‘-’refers to base pair deletion.Lowercase letter‘c’and‘g’in red refer to insertions of‘C’and‘G’,respectively.Num
All of the 126 TKC1.1-LAZY1 T1 plants had a mutation near the PAM site of the target sequence(Table 1).Almost all of the73 TKC1.2-LAZY1 T1 plants were either homozygous or bi-allelic at the LAZY1 locus.However,we found two T1 plants from the#17 T0 plant that had a WT genotype(Table 2),despite that the#17 T0 plant had a lazy phenotype.Our results demonstrated that both TKC1.1 and TKC1.2 vectors were effective for generating targeted editing events.
| 图表编号 | XD0040097800 严禁用于非法目的 |
|---|---|
| 绘制时间 | 2019.03.28 |
| 作者 | HE Yubing、ZHU Min、WANG Lihao、WU Junhua、WANG Qiaoyan、WANG Rongchen、ZHAO Yunde |
| 绘制单位 | National Key Laboratory of Crop Genetic Improvement and National Center of Plant Gene Research (Wuhan), Huazhong Agricultural University、College of Life Science and Technology, Huazhong Agricultural University、College of Plant Science & Technology, Huazho |
| 更多格式 | 高清、无水印(增值服务) |
查看“Table 2.Mutations and segregation analysis of the TKC1.2-LAZY1 T1 plants.”的人还看了
-
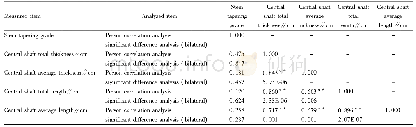
- Table 2 Correlation analysis between stem tapering grade and the growth traits of the central shaft of free spindle-shap
-
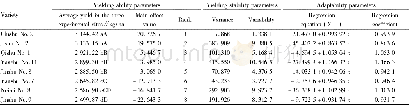
- Table 3 Analysis on yielding ability and stability and adaptability of the test proso millet varieties