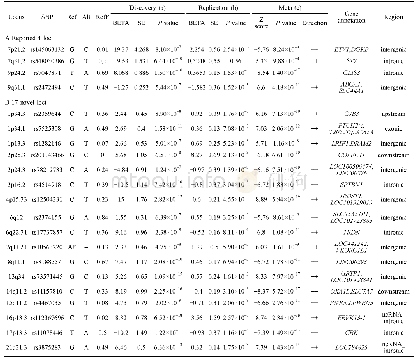
《Table 4 SCP loci and candidate genes significantly associated with SCN resistance of soybean》
 提示:宽带有限、当前游客访问压缩模式
提示:宽带有限、当前游客访问压缩模式
本系列图表出处文件名:随高清版一同展现
《黑龙江省抗胞囊线虫大豆的分子遗传和相关基因挖掘(英文)》
Four genes that were possibly associated with SCN resistance were found(Table 4 and Fig.7):Glyma11g35700.1,which encodes a laccase,is a member of laccase gene family.It is involved in the synthesis of lignin and highly expressed in lignified tissues.When soybean plant is infected by pathogens,Glyma11g35700.1 is largely expressed,which promotes the lignification of the infected part of roots to resist further infection of the pathogens.In addition,it possibly participates in ABA treatment,stress response and lignin catabolism[23].Glyma11g35820.1,encodes alpha-soluble NSF attachment protein 2(ɑ-SNAP2)and mediates membrane fusion.In detail,the adaptor protein of N-ethylmaleimide sensitive fusion proteins recognizes and binds to the receptors V-SNARE(soluble NSF attachment protein receptor residing on the vesicle)on vesicle and t-SNARE on target membranes,initiates the assembly of the fusion complex,which catalyzes the fusion of vesicle with the target membrane.Glyma11g35640.2 encodes DEAD-box ATP-dependent RNA helicase 18,which is involved in the unwinding of RNA duplexes.Glyma11g35651.1 which encodes an autophagy related protein is involved in autophagy of plant cells,and maintains the growth balance in vivo by degrading damaged intracellular organelles,longlived proteins and macromolecular aggregates.
| 图表编号 | XD0014402800 严禁用于非法目的 |
|---|---|
| 绘制时间 | 2018.11.20 |
| 作者 | 魏崃、王伟威、于志远、李云辉、王家军、喻德跃、盖钧镒、刘丽君 |
| 绘制单位 | 南京农业大学大豆研究所、农业部大豆生物学与遗传育种重点实验室、国家大豆改良中心、作物遗传与种质创新国家重点实验室、黑龙江省农业科学院大豆研究所、南京农业大学大豆研究所、农业部大豆生物学与遗传育种重点实验室、国家大豆改良中心、作物遗传与种质创新国家重点实验室、黑龙江省农业科学院大豆研究所、黑龙江省农业科学院大豆研究所、黑龙江省农业科学院大豆研究所、黑龙江省农业科学院大豆研究所、南京农业大学大豆研究所、农业部大豆生物学与遗传育种重点实验室、国家大豆改良中心、作物遗传与种质创新国家重点实验室、南京农业大学大豆研 |
| 更多格式 | 高清、无水印(增值服务) |
查看“Table 4 SCP loci and candidate genes significantly associated with SCN resistance of soybean”的人还看了
-
![表4 盐孢菌属次级代谢产物与对应的生物合成基因簇[16]Table 4 Compounds and their associated BGCs (biosynthetic gene clusters) detected in Salinis](//bookimg.mtoou.info/tubiao/gif/AJSH201805007_37300.gif)
- 表4 盐孢菌属次级代谢产物与对应的生物合成基因簇[16]Table 4 Compounds and their associated BGCs (biosynthetic gene clusters) detected in Salinis
-
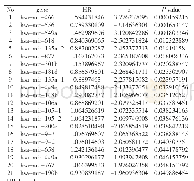
- Table 1.Information on 21 candidate miRNAs associated with OS of breast cancer patients.*Derived from the univariate Cox
-
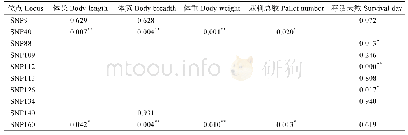
- 表3 SNP位点与体长、体重、体宽、棘刺总数、存活天数相关性分析结果Tab.3 Significant correlation analysis between SNP loci and body length, body weight,
![表4 盐孢菌属次级代谢产物与对应的生物合成基因簇[16]Table 4 Compounds and their associated BGCs (biosynthetic gene clusters) detected in Salinis](http://bookimg.mtoou.info/tubiao/gif/AJSH201805007_37300.gif)