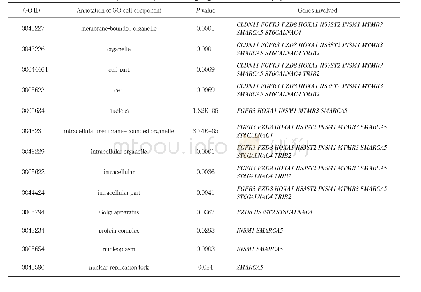
《Table 1 Upregulated and downregulated genes in Schwann cells by microarray analysis》
 提示:宽带有限、当前游客访问压缩模式
提示:宽带有限、当前游客访问压缩模式
本系列图表出处文件名:随高清版一同展现
《Comparative study of microarray and experimental data on Schwann cells in peripheral nerve degeneration and regeneration: big data analysis》
RT-PCR:Reverse transcription-polymerase chain reaction;qPCR:quantitative polymerase chain reaction;ChIP:chromatin immunoprecipitation;WB:western blotting;IHC immunohistochemistry;mRNA:messenger ribonucleic acid;ISH:in situ hybridization.
This study,which is part of a big data analysis,was conducted under the assumption that knowledge of different types of genes and their characteristics and functions(i.e.,whether they are up-or downregulated)will allow for the easy detection of genes that are appropriate for experimental studies on Schwann cells.Additionally,some genes may act differently under different experimental conditions.This method could be a helpful and effective way to detect gene expression in future experimental studies.In conclusion,some genes are highly expressed in microarray studies but weakly expressed in experimental studies.Therefore,a microarray may not be a good tool for Schwann cell studies because the genes may be differently expressed.Conversely,some genes are weakly expressed in microarray studies and highly expressed in experimental studies;these genes are more promising targets for examination in future Schwann cell studies(Table 1).Therefore,microarray analysis could be a good tool for Schwann cell studies.
| 图表编号 | XD0040617500 严禁用于非法目的 |
|---|---|
| 绘制时间 | 2019.06.01 |
| 作者 | Ulfuara Shefa、Junyang Jung |
| 绘制单位 | Department of Biomedical Science,Graduate School,Kyung Hee University、Department of Biomedical Science,Graduate School,Kyung Hee University、Department of Anatomy and Neurobiology,College of Medicine,Kyung Hee University |
| 更多格式 | 高清、无水印(增值服务) |