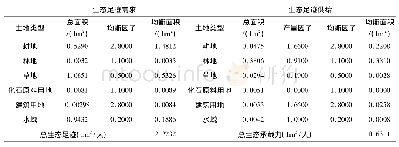
《Table 3 Summary of the susceptibility genes and SNPs in glioma GWAS》
*Intronic variants
In the post-genome sequenc ing era,over 10million SNPs are now documented,coupled with the development of highly efficient analytical platforms and techniques are allowing genome-wide association analysis(GWAS)to be conducted in a cost-effective manner(Table 3).By adopting this approach,important progress has been made to understand host susceptibility to brain tumor.Using GWAS,5 risk loci for glioma have been identified in the first glioma GWAS including1 878 glioma patients and 3 670 controls,and validated in 2 545 glioma patients and 2 953 controls:5p15.33TERT(telomerase reverse transcriptase),8q24.21CCDC26(coiled-coil domain containing 26),9p21.3CDKN2A-CDKN2B(cyclin dependent kinase inhibitor),20q13.33 RTEL1(regulator of telomere elongation helicase 1),and 11q23.3 PHLDB1(pleckstrin homology like domain family B member 1)[30].Wrensch et al[31]c o n t r i b u te d ad d i t i o n a l e v i d e n c e i m p l i c at i ng t h e CDKN2A-CDKN2B,RTEL1,and TERT variants in highgrade gliomas.Another GWAS identified 2 additional genetic variants[32]:7p11.2 EGFR(epidermal growth factor receptor)and 7q36.1 XRCC2(X-ray repair cross complementing 2).Recently,a Meta-analysis of existing GWAS and 2 new GWAS,with total 12 496 glioma cases and 18 190 controls,has been performed by the Glioma International Case Control Consortium[33].This study has been identified 5 new loci for GBM at 1p31.3 RAVER2(ribonucleoprotein,PTB binding 2),11q14.1 RPS28P7(ribosomal protein S28 pseudogene 7),16p13.3 MPG(N-methylpurine DNA glycosylase),16q12.1 HEATR3(HEAT Repeat Containing 3),and 22q13.1 SLC16A8(solute carrier family 16 member 8),as well as 8 loci for non-GBM tumors at 1q32.1 MDM4(mouse double minute 4,human homolog;P53-binding protein),1q44AKT3(AKT serine/threonine kinase 3),2q33.3 IDH1(isocitrate dehydrogenase 1,cytosolic),3p14.1 LRIG1(leucine rich repeats and immunoglobulin like domains1),10q24.33 OBFC1(oligonucleotide/oligosaccharidebinding fold-containing protein 1),11q21 MAML2(mastermind like transcriptional coactivator 2),14q12AKAP6(A-kinase anchoring protein 6),and 16p13.3LMF1(lipase maturation factor 1).These data substantiate that genetic susceptibility to GBM and non-GBM tumors are highly distinct,which likely reflects different etiologies.In addition,inherited variants of LIG4(DNA ligase 4),BTBD2(BTB domain containing 2),HMGA2(high mobility group AT-hook 2),and RTEL1 genes which involved in the double-strand break repair pathway are also associated with GBM survival[34]and neurocognitive function[35].
| 图表编号 | XD0026936200 严禁用于非法目的 |
|---|---|
| 绘制时间 | 2018.04.01 |
| 作者 | 李学军、曹航、LIU Yanhong |
| 绘制单位 | 中南大学湘雅医院神经外科、中南大学湘雅医院颅底外科与神经肿瘤研究中心、中南大学湘雅医院神经外科、中南大学湘雅医院颅底外科与神经肿瘤研究中心、贝勒医学院丹L.邓肯综合癌症研究中心医学系 |
| 更多格式 | 高清、无水印(增值服务) |
查看“Table 3 Summary of the susceptibility genes and SNPs in glioma GWAS”的人还看了
-
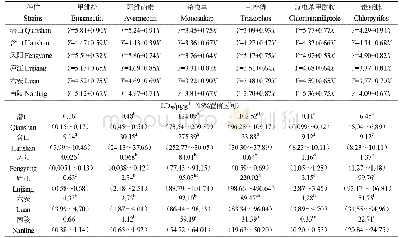
- 表3 安徽二化螟不同地理种群对主要杀虫剂的敏感性测定 (2016) Table 3 The insecticide susceptibility of Chilo suppressalis in Anhui Province (2016)
-
- Table 1 Metadata summary of the“Dataset of fruit types and seed dispersal modes of plants in five com-munities in Shilin
-

- Table 4.Correlation between expression level of ROCK1 gene and the therapeutic efficacy in ALL patients (n, %)
 提示:宽带有限、当前游客访问压缩模式
提示:宽带有限、当前游客访问压缩模式