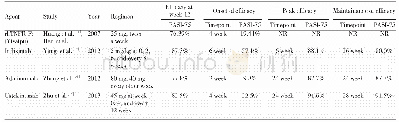
《Table 1Basic data information of four sections in the CCGD-ESCC》
 提示:宽带有限、当前游客访问压缩模式
提示:宽带有限、当前游客访问压缩模式
本系列图表出处文件名:随高清版一同展现
《CCGD-ESCC: A Comprehensive Database for Genetic Variants Associated with Esophageal Squamous Cell Carcinoma in Chinese Population》
Note:GWAS,genome-wide association study;eQTL,expression quantitative trait loci;ESCC,esophageal squamous-cell carcinoma;WGS,whole-genome sequencing;WES,whole-exome sequencing;SNP,single nucleotide polymorphism;SNV,single nucleotide variation.
To build the CCGD-ESCC database,we integrated multiple genetic association results and set up four sections.These include(1)GWAS of 16,544 SNPs in 2022 ESCC cases and2039 controls;(2)survival GWAS of 1652 SNPs in 1006 ESCC patients;(3)eQTLs from 94 ESCC patients with both WGS and RNA-seq data in tumor and paired normal tissues;and(4)8833 single nucleotide variations(SNVs)/indels in the protein-coding regions from 675 ESCC patients with WGS or WES data and their associations with survival time in ESCC patients(Table 1).The criteria of enrollment of study subjects were described in the previous publications[6,7,9,14,22].Genetic variants were annotated using the ANNOVAR with the reference cluster identifier(rsID)from the NCBI dbSNP build 146[23].
| 图表编号 | XD0021323300 严禁用于非法目的 |
|---|---|
| 绘制时间 | 2018.08.01 |
| 作者 | Linna Peng、Sijin Cheng、Yuan Lin、Qionghua Cui、Yingying Luo、Jiahui Chu、Mingming Shao、Wenyi Fan、Yamei Chen、Ai Lin、Yiyi Xi、Yanxia Sun、Lei Zhang、Chao Zhang、Wen Tan、Ge Gao、Chen Wu、Dongxin Lin |
| 绘制单位 | Department of Etiology and Carcinogenesis, National Cancer Center、Cancer Hospital, Chinese Academy of Medical Sciences and Peking Union Medical College、State Key Laboratory of Protein and Plant Gene Research, School of Life Sciences, Center for Bioinforma |
| 更多格式 | 高清、无水印(增值服务) |