《Table 2 The comparison analysis of amino acid sequences using DNAMAN》
 提示:宽带有限、当前游客访问压缩模式
提示:宽带有限、当前游客访问压缩模式
本系列图表出处文件名:随高清版一同展现
《Bioinformatics Analysis on Pathogenic Genes of Botrytis cinerea Pers.》
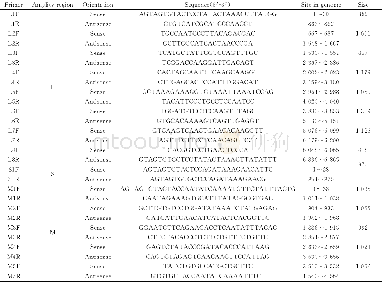
The protein sequences of Aac were compared with corresponding amino acid sequences of pathogenic genes of other plant pathogens.It could be seen from Table 2 that yp_968837 and AAZ34500,yp_968846 and AAW73346,yp_968848 and AAF61285,yp_96929 and AAZ356963,yp_970484 and CEL40510,and yp_970742 and CAO95583 were aligned.The homology values were not high,but the similar residues all reached one half or more,and except that protein yp_970484 had a higher value,the gap values were all lower than one half.Comprehensively,the three pairs of proteins with higher matching degrees were yp_968848 and AAF61285,yp_969269 and AAZ35693,and yp_968837 and AAZ34500,indicating that the similarity between the functions of each pair was higher.Because these pathogenic genes of other plant bacterial pathogens are all from the hrp gene cluster of type III secretion system,these proteins were also predicted to be coded by hrp gene cluster.
| 图表编号 | XD00183011400 严禁用于非法目的 |
|---|---|
| 绘制时间 | 2018.02.01 |
| 作者 | Qiang ZHANG、Wenting DAI、Xinwen JIN、Xuhui WANG |
| 绘制单位 | Xinjiang Agricultural Reclamation Academy of Science、Xinjiang Agricultural Reclamation Academy of Science、Xinjiang Agricultural Reclamation Academy of Science、Organisms Energy Research Institute in Xin Jiang Academy of Agriculture Science |
| 更多格式 | 高清、无水印(增值服务) |
查看“Table 2 The comparison analysis of amino acid sequences using DNAMAN”的人还看了
-
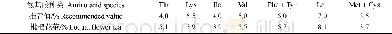
- 表2 枇杷花茶中各种必需氨基酸所占比值与WHO/FAO推荐氨基酸模式谱的比较Table 2 The comparison of various essential amino acids and WHO/FAO recommended am