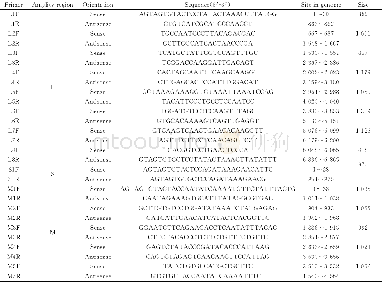
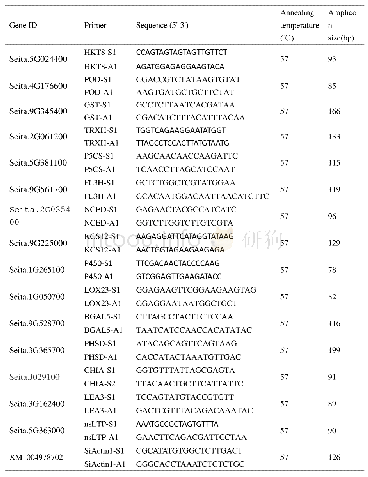
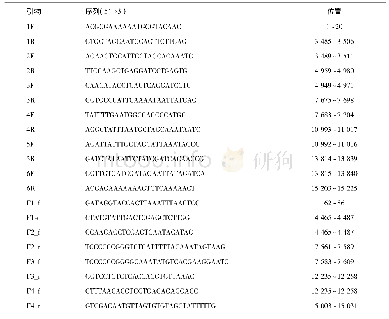
《Table 1.List of primer used for the amplification of the four pierid butterfly mitogenome.》
 提示:宽带有限、当前游客访问压缩模式
提示:宽带有限、当前游客访问压缩模式
本系列图表出处文件名:随高清版一同展现
《Mitochondrial genomes of four pierid butterfly species(Lepidoptera: Pieridae) with assessments about Pieridae phylogeny upon multiple mitogenomic datasets》
*Annealing temperature
Total genomic DNA was extracted from leg muscle of each adult individual by the glass powder method with minor modifications(Hao et al.,2005).Three partial fragments of COI,ND4,and Cytb were amplified by using standard short primers from Simon et al.(1994),other primers for the amplications of long and short fragments were designed by Primer Premier 5.0(Singh et al.,1998)through multiple sequence alignments of butterfly mitogemomes using Clustal X1.8(Thompson et al.,1997).All the primers were synthesized by Sangon Biotechnology Co.Ltd.(Shanghai,China)(Table 1) .Long PCR amplification reactions were performed under the following conditions:an initial denaturation at 95°C for 5 min;30 cycles of denaturation(denaturing at 95°C for 50 s,annealing at 50°–55°C for 50 s,extension at 68°C for 150 s),and a final extension step at 68°C for 10 min.The PCR products were separated by 1.2%agarose gel electrophoresis,purified by using a 3S Spin PCR Product Purification Kit(Sangon Biotechnology Co.Ltd.,Shanghai)and sequenced on an ABI-377automatic DNA sequences.For each long PCR product,the complete,double-stranded sequence was determined by the“primer walking”method.
| 图表编号 | XD0016211300 严禁用于非法目的 |
|---|---|
| 绘制时间 | 2018.10.01 |
| 作者 | Lan Nie、Yunliang Wang、Dunyuan Huang、Ruisong Tao、Chengyong Su、Jiasheng Hao、Chaodong Zhu |
| 绘制单位 | Laboratory of Molecular Evolution and Biodiversity, College of Life Sciences, Anhui Normal University、Laboratory of Molecular Evolution and Biodiversity, College of Life Sciences, Anhui Normal University、College of Life Sciences, Chongqing Normal Universi |
| 更多格式 | 高清、无水印(增值服务) |