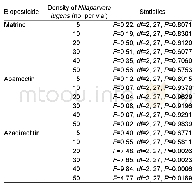
《Table 2–Summary statistics from bioinformatic analysis and wet-lab validation.》
 提示:宽带有限、当前游客访问压缩模式
提示:宽带有限、当前游客访问压缩模式
本系列图表出处文件名:随高清版一同展现
《"Identification, development, and application of cross-species intron-spanning markers in lentil(Lens culinaris Medik.)"》
A total of 1703 ISMs were developed in lentil using a cross-species mapping-based approach.For the identification of ISMs,16,279 EST sequences of L.culinaris from were mapped onto the M.truncatula genome(Table 2).Rep Base libraries and L.culinaris ESTs were aligned which resulted in a total of 25,717Gene Seqer alignments.These were further curated to identify1703 ISMs(Table S2).Among these,a set of 105 primer pairs were used for experimental validation,which resulted in successful amplification of 54 primer pairs(51%)on lentil genomic DNA.
| 图表编号 | XD0012324200 严禁用于非法目的 |
|---|---|
| 绘制时间 | 2018.06.01 |
| 作者 | Debjyoti Sen Gupta、Jitendra Kumar、SunANDa Gupta、Sonali Dubey、Priyanka Gupta、Narendra Pratap Singh、Gaurav Sablok |
| 绘制单位 | Division of Crop Improvement,ICAR-Institute of Pulses Research、Division of Crop Improvement,ICAR-Institute of Pulses Research、Division of Crop Improvement,ICAR-Institute of Pulses Research、Division of Crop Improvement,ICAR-Institute of Pulses Research、Div |
| 更多格式 | 高清、无水印(增值服务) |
查看“Table 2–Summary statistics from bioinformatic analysis and wet-lab validation.”的人还看了
-
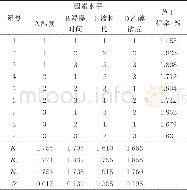
- 表2 芦丁得率正交实验结果与分析Table 2 Results and analysis of orthogonal test of extract-ing rutin from jujube with ethanol
-
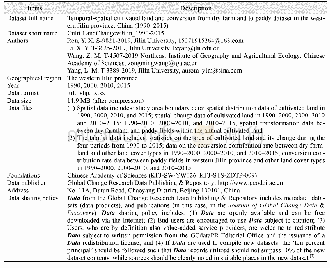
- Table 1 Metadata summary of the“Temporal-spatial cultivated land and conversion from dry farmland to paddy dataset in th
-
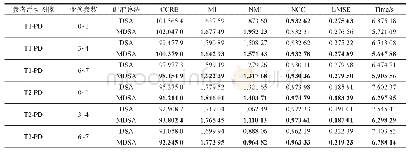
- Table 2 Statistical analysis of DSA and MDSA in 6 multi-spectral medical image registrations表2 6组多谱医学图像配准中DSA和MDSA的性能统计
-
- Table 6 Summary of major risk factors for perforation and bleeding related to colonoscopy from recent studies with sampl