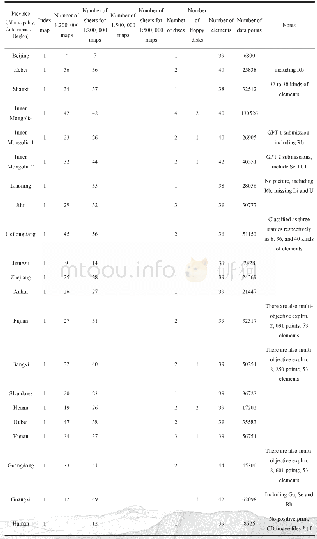
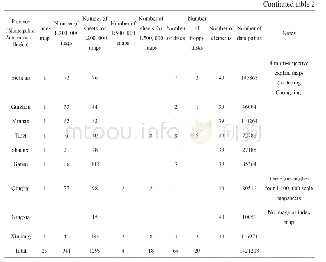
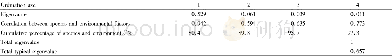
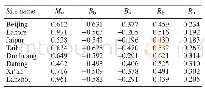
《Table 2–Summary statistics of the simple sequence repeat (SSR) motifs designed with primers and dis
 提示:宽带有限、当前游客访问压缩模式
提示:宽带有限、当前游客访问压缩模式
本系列图表出处文件名:随高清版一同展现
《Development and validation of simple sequence repeat markers from Arachis hypogaea transcript sequences》
To validate the identified novel SSR markers,we attempted to amplify the predicted SSRs via PCR.A total of 210 primer pairs(Table S2)were randomly selected for validation using DNA from 24 A.hypogaea varieties(Table S1).The numbers of these selected SSRs with di-,tri-,and tetranucleotide repeats were 35,153,and 22,respectively.Among the 210 primer pairs,191(90.95%)were able to amplify genomic DNA and the containing SSRs with di-,tri-,and tetranucleotide repeats were 32(16.75%),140(73.30%),and 19(9.95%),respectively,whereas the remaining 19 primer pairs failed to generate PCR products at the same annealing temperatures(Table S2).Most of the selected markers appeared as single alleles in all 24 A.hypogaea genotypes,except for a few multilocus SSRs,suggesting that these novel SSR markers possess a specific amplification in A.hypogaea.
| 图表编号 | XD0012317900 严禁用于非法目的 |
|---|---|
| 绘制时间 | 2018.04.01 |
| 作者 | Houmiao Wang、Yong Lei、Liying Yan、Liyun Wan、Yan Cai、Zefeng Yang、Jianwei Lv、Xiaojie Zhang、Chenwu Xu、Boshou Liao |
| 绘制单位 | Key Laboratory of Oil Crop Biology of the Ministry of Agriculture, CAAS-ICRISAT Joint Laboratory for Groundnut Aflatoxin Management, Oil Crops Research Institute of Chinese Academy of Agricultural Sciences、Jiangsu Key Laboratory of Crop Genetics and Physi |
| 更多格式 | 高清、无水印(增值服务) |
查看“Table 2–Summary statistics of the simple sequence repeat (SSR) motifs designed with primers and distribution of new SSRs”的人还看了
-

- 表1 2:Descriptive Statistics of the Scores at Grammatical Level Formulaic Sequences in the Delayed Post Test Dependent Va