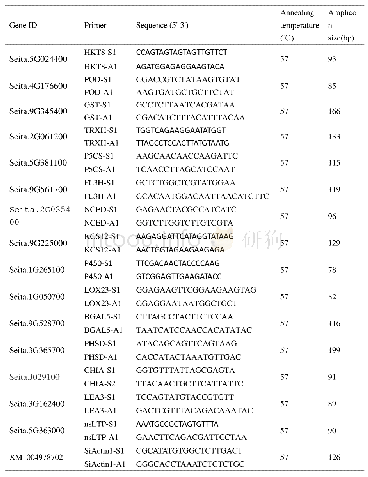
《Table 1.Nucleotide sequences used in construction of fusion vector with chloroplast transit peptide
 提示:宽带有限、当前游客访问压缩模式
提示:宽带有限、当前游客访问压缩模式
本系列图表出处文件名:随高清版一同展现
《Functional Analysis of Three Rice Chloroplast Transit Peptides》
To construct the p35S::CTPTRXz-GFP vector,we generated a DNA fragment encoding the N-terminal56 amino acids of TRXz by PCR using p35S::CTPTRXz-GFP primer(Table 1)from the NPB cDNA.The PCR procedure was as follows:Denaturation at94oC for 4 min,followed by 32 cycles of 98oC for 10s,annealing at 55oC for 30 s,extension at 68oC for30 s,and a final extension step at 68oC for 10 min.The PCR products were analyzed on 2%agarose gels.We used the Wizard SV Gel and PCR Clean-Up System(Promega Biotech,USA)to extract and purify the target DNA fragment.The purified DNA fragment encoding the N-terminal 56 amino acids of TRXz was fused with the purified p35S::GFP fragment using a ClonExpress II One Step Cloning Kit(Vazyme Biotech,Nanjing,China)by homologous recombination.The ligation reaction was conducted for 30 min at37oC in a 20μL reaction,consisting of 2μL PCR product,2μL purified p35S::GFP fragment,4μL of5×CEII buffer,2μL Exnase II and 10μL sterile water.After transformation into Escherichia coli(DH5α),the positive clones were identified by PCR using the s65t-1F(GAGGACAGGCTTCTTGAG)and s65tR(GGTGGTGCAGATGAACTT)primers and sent to Hangzhou Tsingke Biotech Company(Hangzhou,China)for sequencing verification using the s65t-1F and s65tR primers.The other CTP and GFP fusion protein vectors(p35S::CTPHSA1-GFP and p35S::36CTPFLN1-GFP)were constructed and verified as described above using primers listed in Table 1.
| 图表编号 | XD0040096400 严禁用于非法目的 |
|---|---|
| 绘制时间 | 2019.01.28 |
| 作者 | HE Lei、CHEN Guang、ZHANG Sen、QIU Zhennan、HU Jiang、ZENG Dali、ZHANG Guangheng、DONG Guojun、GAO Zhenyu、REN Deyong、SHEN Lan、GUO Longbiao、QIAN Qian、ZHU Li |
| 绘制单位 | State Key Laboratory of Rice Biology, China National Rice Research Institute、State Key Laboratory of Rice Biology, China National Rice Research Institute、State Key Laboratory of Rice Biology, China National Rice Research Institute、State Key Laboratory of |
| 更多格式 | 高清、无水印(增值服务) |