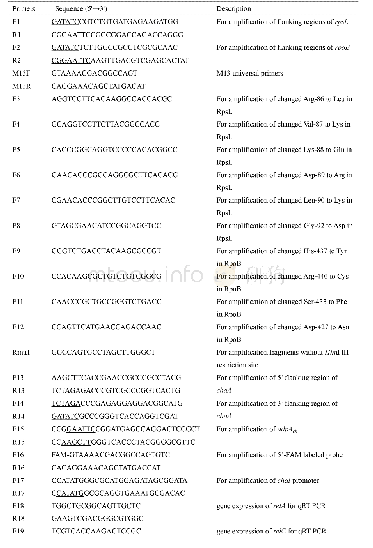
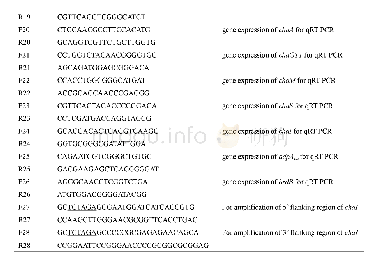
《Table S1 DNA oligonucleotide sequences used in this study》
As illustrated in Fig.1A,the SSS is in silico designed as the hairpin structure(Ⅰ)consisting of five domains,in which domains b and b*,c and c*,d and d*hybridize together forming the stem of hairpin structure.A 20 nt single-stranded DNA trigger consisting of domains e*and d*can bind to the“toehold”region of SSS at the domain e of SSS.And the domain d*of trigger can further hybridize with domain d forcing SSS to become a second hairpin structure(Ⅱ),in which 3'end of domain c forms a stem structure for further DNA amplification.When Taq DNA polymerase was supplemented,DNA extended from domain c,eventually forming structure(Ⅲ).During the conversion process from structure(Ⅱ)to structure(Ⅲ),the trigger will be removed by the highly processive 5'-3'exonuclease activity of Taq DNA polymerase(Taq DNAP)and the whole oneway reaction will be terminated.The dynamic structure change was detected by PAGE analysis.In the presence of SSS and Taq DNAP,the band of SSS(63 nt)migrated down.We suspected the reason was that the SSS was degraded by Taq DNA polymerase.In the presence of trigger molecule(20 nt),the band of structure(Ⅰ)migrated upward(Fig.1B),indicating that the structure(Ⅱ)has been formed by a specific hybridization between trigger and toehold region of SSS.After the Taq DNAP was added to SSS,the band of structure(Ⅱ)moved upward further and indicated that structure(Ⅲ)has been formed with the action of Taq DNAP(Fig.1B).We also tested two complete in silico designed SSS sequences,Probe-1 and Probe-2(Supplementary Table S1),which effectively reacted with trigger molecule(Supplementary Fig.S1).
| 图表编号 | XD0094291600 严禁用于非法目的 |
|---|---|
| 绘制时间 | 2019.10.20 |
| 作者 | 胡亚赛、郜艳敏、郝敏、齐浩 |
| 绘制单位 | 天津大学化工学院系统生物工程教育部重点实验室、天津大学化工学院系统生物工程教育部重点实验室、天津大学化工学院系统生物工程教育部重点实验室、天津大学化工学院系统生物工程教育部重点实验室 |
| 更多格式 | 高清、无水印(增值服务) |
 提示:宽带有限、当前游客访问压缩模式
提示:宽带有限、当前游客访问压缩模式