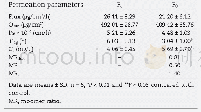
《Table 1 Pioneering studies in application of gut microbiome approaches in conservation biologya)》
 提示:宽带有限、当前游客访问压缩模式
提示:宽带有限、当前游客访问压缩模式
本系列图表出处文件名:随高清版一同展现
《Conservation metagenomics: a new branch of conservation biology》
a) 1,T means targeted sequencing(such as 16S rRNA);S means metagenomic shotgun sequencing;M means metabolomics methods.R means metatranscriptomic methods.P means(real time)PCR.2,D means that the research focuses on the diversity of microbiome.F means
The ongoing development in metagenomics technologies as well as the introduction of non-invasive sampling methods enables deep access to the gut microbiome of wild animals.Some pioneering research focused on the evolutionary relationship between phylogeny and diet of hosts and composition/function of gut microbiome in mammals(Ley et al.,2008;Muegge et al.,2011).Following these global patterns of mammals,some intensive work on individual species has also been reported,including the tammar wallaby(Pope et al.,2010)and the giant panda(Zhu et al.,2011).The potential role of gut microbiome in conservation biology has been gradually revealed,including host health and disease,habitat degradation and nutrition utilization of endangered animals(Stumpf et al.,2016).Great achievements have been made in this field during the past decade(Ding et al.,2017;Table 1).
| 图表编号 | XD0034728000 严禁用于非法目的 |
|---|---|
| 绘制时间 | 2019.02.05 |
| 作者 | Fuwen Wei、Qi Wu、Yibo Hu、Guangping Huang、Yonggang Nie、Li Yan |
| 绘制单位 | CAS Key Laboratory of Animal Ecology and Conservation Biology, Institute of Zoology, Chinese Academy of Sciences、University of Chinese Academy of Sciences、Center for Excellence in Animal Evolution and Genetics, Chinese Academy of Sciences、CAS Key Laborato |
| 更多格式 | 高清、无水印(增值服务) |
查看“Table 1 Pioneering studies in application of gut microbiome approaches in conservation biologya)”的人还看了
-
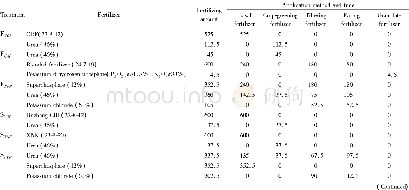
- Table 2 Quantity and application method of fertilizer application in the early and late rice cropping fields and LAD in
-
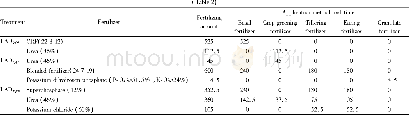
- Table 2 Quantity and application method of fertilizer application in the early and late rice cropping fields and LAD in