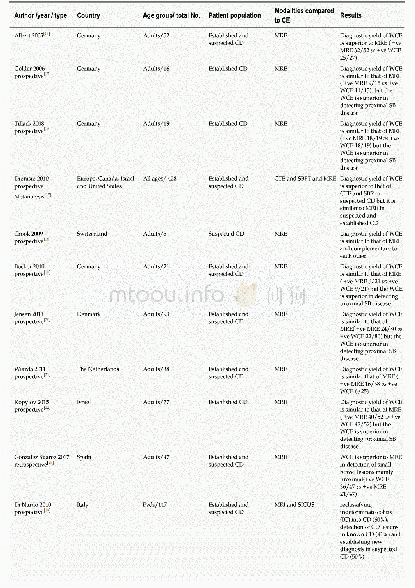
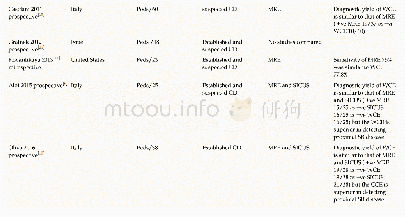
《Table 1 Summary of mutations in MARV/Ang-GA, compared to MARV/Ang》
 提示:宽带有限、当前游客访问压缩模式
提示:宽带有限、当前游客访问压缩模式
本系列图表出处文件名:随高清版一同展现
《Development and characterization of a guinea pig model for Marburg virus》
N/A:not applicable.
Since the virus passaging by Cross et al.(2015)was performed completely independently from our study,this provided a rare opportunity to compare viral genome sequences between their guinea pig-adapted virus versus our MARV/Ang-GA.The purpose was to see if any common mutations exist between the two adapted viruses,which may give indications of the relative importance of each mutation.To our surprise,the two viruses were remarkably similar.All mutations in the coding and non-coding regions were identical,save for the N242S mutation in VP24 described in our study that was not present in their variant,and a mutation in a non-coding region(base pair 19 105)that was described in their study but undetected in our viral stock(Table 1).This suggested that the common mutations between our two viral stocks were the critical changes needed to attain virulence in guinea pig;however,it should be noted that some of the mutations may have been introduced during the preparation of the virus stock in Vero E6 cells,as previously demonstrated with a mouse-adapted MARV/Ang(Wei et al.,2017).It will be interesting to sequence the virus from the last in vivo passage from the guinea pigs and compare with the sequence of our viral stock.Additionally,in-depth studies using the reverse genetics system for MARV will be needed to elucidate the impact of each mutation.
| 图表编号 | XD0016284600 严禁用于非法目的 |
|---|---|
| 绘制时间 | 2018.01.18 |
| 作者 | Gary Wong、Wen-Guang Cao、Shi-Hua He、Zi-Rui Zhang、Wen-Jun Zhu、Estella Moffat、Hideki Ebihara、Carissa Embury-Hyatt、Xiang-Guo Qiu |
| 绘制单位 | Special Pathogens Program, National Microbiology Laboratory, Public Health Agency of Canada、Shenzhen Key Laboratory of Pathogen and Immunity, State Key Discipline of Infectious Disease, Shenzhen Third People's Hospital、Key Laboratory of Pathogenic Microb |
| 更多格式 | 高清、无水印(增值服务) |