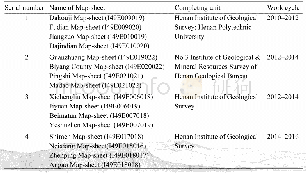
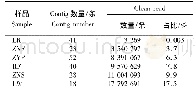
《Table 3 Results of clean reads mapping to Glymacine max genome》
 提示:宽带有限、当前游客访问压缩模式
提示:宽带有限、当前游客访问压缩模式
本系列图表出处文件名:随高清版一同展现
《Comparative Transcriptome Profiling of Glycine soja Roots Under Salinity and Alkalinity Stresses Using RNA-seq》
The total RNA isolated from the root samples were used for RNA-seq analysis.RNA was reversetranscribed to c DNA by using Super Script TMⅢReverse Transcriptase kit(Invitrogen,Cal,USA).The quantitative real-time PCR(q RT-PCR)was performed with each c DNA template using SYBR Premix ExTaq™;ⅡMix(Ta Ka Ra,Shiga,Japan)using an ABI7500 sequence detection system(Applied Biosystems,Carlsbad,CA).GAPDH(accession#DQ355800)was used to normalize all the estimated values.The primers for q RT-PCR for 14 DEGs under salt and alkali stresses were designed using Primer Premier 5.0(http://www.premierbiosoft.com/primerdesign/)software and listed in Table 1a and Table 1b.The primers for q RT-PCR of eight MYB TFs under salt and alkali stresses are listed in Table 2a and Table 2b.Relative expression levels of DEGs were calculated using the2-ΔΔCT method.Three independent biological replicates were used for q RT-PCR in each of the three technical replicates.
| 图表编号 | XD0014156000 严禁用于非法目的 |
|---|---|
| 绘制时间 | 2018.09.25 |
| 作者 | Zhu Yan-ming、Li Ji-na、DuanMu Hui-zi、Yin kui-de、Cheng Shu-fei、Chen Chao、Cao Lei、Duan Xiang-bo、Chen Ran-ran |
| 绘制单位 | College of Life Sciences, Northeast Agricultural University、College of Life Sciences, Northeast Agricultural University、College of Life Sciences, Heilongjiang University、College of Life Sciences and Technology, Heilongjiang Bayi Agricultural University、Co |
| 更多格式 | 高清、无水印(增值服务) |