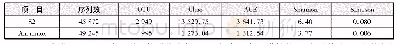《Table 1–Microbial species richness and diversity indices in UBAF-only and ICME–UBAF coupled (C-UBAF
 提示:宽带有限、当前游客访问压缩模式
提示:宽带有限、当前游客访问压缩模式
本系列图表出处文件名:随高清版一同展现
《Enhanced removal of organic matter and typical disinfection byproduct precursors in combined iron–carbon micro electrolysis-UBAF process for drinking water pre-treatment》
16S rRNA sequencing was employed in order to determine the richness and diversity of bacterial communities in the two systems.These sequences were subsequently clustered into operational taxonomic units(OTUs).OTUs refer to clusters of(uncultivated or unknown)organisms,grouped by DNA sequence similarity of a specific taxonomic marker gene such as 16S rRNA(Revetta et al.,2010).Results suggested that the OTUs captured around 98%of species of all the samples,and therefore they were representative enough for the analysis.The ACE index and Chao1 index are quantitatively estimated to reflect the richness of microbial species larger ACE and Chao1 indexes correspond to higher richness of species in a bacterial community.The Shannon index as well as the Simpson index,is a quantitative measure reflecting the diversity of microbial species:higher Shannon index and lower Simpson index mean higher community diversity(Zhao et al.,2016).According to Table 1,the Shannon index of ICME–UBAF was higher and the Simpson index was lower than that of the UBAF alone,indicating the higher diversity of microbial species in ICME–UBAF.The higher Chao1 index and higher ACE index indicated that the ICME–UBAF had higher richness of microbial species.The above results showed that the richness and diversity of the microbial community increased in the combined ICME–UBAF due tothe changes in the physicochemical properties of the feed water.
| 图表编号 | XD0033528000 严禁用于非法目的 |
|---|---|
| 绘制时间 | 2019.04.15 |
| 作者 | Yinghan Chen、Tao Lin、Wei Chen |
| 绘制单位 | Ministry of Education Key Laboratory of Integrated Regulation and Resource Development on Shallow Lakes,Hohai University、College of Environment,Hohai University、Ministry of Education Key Laboratory of Integrated Regulation and Resource Development on Shal |
| 更多格式 | 高清、无水印(增值服务) |
查看“Table 1–Microbial species richness and diversity indices in UBAF-only and ICME–UBAF coupled (C-UBAF) systems.”的人还看了
-
- Table 1 Individual densities and species richness of soil fauna during litter decomposition of bamboo(Fargesia spathacea
-
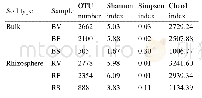
- Table 2.Alpha diversity indices and numbers of OTUs in rhizosphere and bulk soil microbial communities in different grow
-
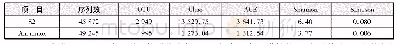
- 表3 SBBR反应器中Anammox菌丰度变化情况Tab.3 Variations of richness and diversity estimators of microbial communities in SBBR reactor