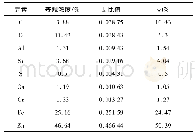
《Table 3–Chromosome locations of P-genome-specific SNPs in wheat-A.cristatum addition lines.》
 提示:宽带有限、当前游客访问压缩模式
提示:宽带有限、当前游客访问压缩模式
本系列图表出处文件名:随高清版一同展现
《Development of P genome-specific SNPs and their application in tracing Agropyron cristatum introgressions in common wheat》
Eighteen genetically diverse common wheat accessions and A.cristatum(L.)accession Z559 were used for screening P genome-specific SNP markers.A.cristatum(L.)accession Z559(2n=4x=28,PPPP)served as the male donor parent in the original cross between wheat and A.cristatum.The 18accessions included Fukuhokomugi,the female parent used in the original wide cross,Chinese Spring,Gaocheng 8901,Xiaoyan 6,Zhoumai 18,Jimai 22,Shi 4185,Yumai 18,Lumai 14,Zhengmai 9023,Han 4589,Dongshi 1,and Zhongmai 175 from China,and Jagger,Hi-Line,McGuire,CMH83.605 and Lovin 10from other countries(Table S1).Seventeen wheat-A.cristatum disomic addition lines were used to determine the chromosome locations of the P genome-specific SNPs.These addition lines included two 1P addition lines,three 2P addition lines,one 1P and 2P addition line,two 4P addition lines,one 5P addition line,four 6P addition lines and four 7P addition lines(Table 1).A set of 19 wheat-A.cristatum 6P translocation lines(Table S2)[42]were used to physically map chromosome6P-specific SNPs.Eighty-nine wheat-A.cristatum derivatives were used to detect whether they carried P chromosomal fragments or functional genes.
| 图表编号 | XD0029304100 严禁用于非法目的 |
|---|---|
| 绘制时间 | 2019.04.01 |
| 作者 | Huihui Ma、Jinpeng Zhang、Jing Zhang、Shenghui Zhou、Haiming Han、Weihua Liu、Xinming Yang、Xiuquan Li、Lihui Li |
| 绘制单位 | Institute of Crop Sciences, Chinese Academy of Agricultural Sciences、Institute of Crop Sciences, Chinese Academy of Agricultural Sciences、Institute of Crop Sciences, Chinese Academy of Agricultural Sciences、Institute of Crop Sciences, Chinese Academy of A |
| 更多格式 | 高清、无水印(增值服务) |