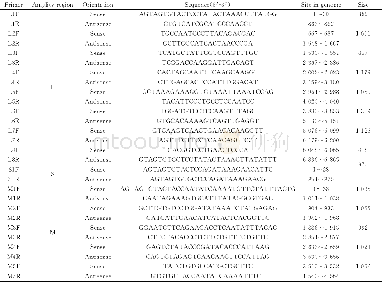
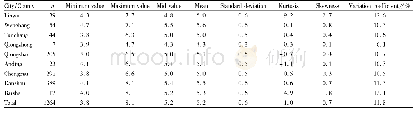
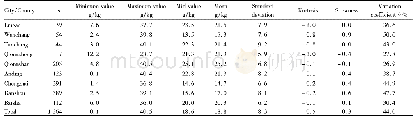
《Table 2 Assembly information of Hainan Partridge genome》
 提示:宽带有限、当前游客访问压缩模式
提示:宽带有限、当前游客访问压缩模式
本系列图表出处文件名:随高清版一同展现
《First complete genome sequence in Arborophila and comparative genomics reveals the evolutionary adaptation of Hainan Partridge(Arborophila ardens)》
The GC content of the Hainan Partridge genome was approximately 42.17%,similar to other bird species such as the Ground Tit,Red Junglefowl and Zebra Finch(Cai et al.2013).The repeat sequences are about 9.19%(96.04 Mb)including long interspersed nuclear elements(LINEs,6.70%),long terminal repeats(LTRs,1.27%),short interspersed nuclear elements(SINEs,0.06%),and DNA transposons 1.14%(Additional file 1:Table S3).A total of 17,376 PCGs in Hainan Partridge genome were predicted and most(92.03%)of them were well supported by public protein databases(TrEMBL,Swissprot,Nr,InterPro,GO and KEGG)(Fig.2b,Additional file 1:Table S4) .The average length of genes and coding sequences were 24,359 bp and 1689 bp with an average of10 exons per gene.
| 图表编号 | XD0047412900 严禁用于非法目的 |
|---|---|
| 绘制时间 | 2019.03.01 |
| 作者 | Chuang Zhou、Shuai Zheng、Xue Jiang、Wei Liang、Megan Price、Zhenxin Fan、Yang Meng、Bisong Yue |
| 绘制单位 | Key Laboratory of Bioresources and Ecoenvironment(Ministry of Education),College of Life Sciences,Sichuan University、Key Laboratory of Bioresources and Ecoenvironment(Ministry of Education),College of Life Sciences,Sichuan University、Sichuan Key Laborator |
| 更多格式 | 高清、无水印(增值服务) |