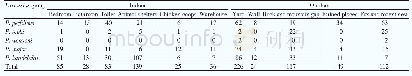
《Table 1.Genetic diversity of Enhalus acoroides collected in six different seagrass beds in both the
 提示:宽带有限、当前游客访问压缩模式
提示:宽带有限、当前游客访问压缩模式
本系列图表出处文件名:随高清版一同展现
《"Assessment by microsatellite analysis of genetic diversity and population structure of Enhalus acoroides from the coast of Khanh Hoa Province, Vietnam"》
Note:Cr represents Cymodocea rotundata,Cs Cymodocea serrulata,Ea Enhalus acoroides,Ho Halophila ovalis,Hu Halodule uninervis,Th Thalassia hemprichii,*Dominant species,HO observed heterozygosity,HE expected heterozygosity,A allele richness,and FIS
Young leaves of each individual plant were ground in a bead mill(22 Hz,2 min)(Retsch,Technology GmbH,Haan,Germany) ,and 100 mg of the finely powdered plant material was used for DNA extraction.DNA was extracted by the Plant Nucleospin II Kit(Macherey&Nagel,Düren,Germany),following manufacturer’s instruction with a few modifications according to Lucas et al.(2012).DNA quality was checked on agarose gels stained with ethidium bromide.DNA concentration was quantified using a microplate reader with micro-volume plates(Synergy Mx MultiMode,BioTek,Germany).Sixty individuals collected from six populations along the coast of the Khanh Hoa Province were used for the analysis.Details of sample size,locations and coordinates are shown in Table 1.Among 10 primer pairs suggested by Nakajima et al.(2012),we used five primer pairs resulting in highly polymorphic bands(Eaco_009,Eaco_050,Eaco_051,Eaco_054,and Eaco_055)for PCR.Thirty ng of template DNA was used in each 15μL PCR including 1x Williams buffer(10 mmol/L Tris/HCl pH 8.3,50 mmol/L KCl,2 mmol/L MgCl2,0.001%gelatin),0.2 mmol/L dNTPs,1 U Dream Taq DNA polymerase(Thermo Fisher Scientific,Dreieich,Germany),and 1 pmol primer each.The PCR was performed in a PTC 200 thermocycler(Biozym-Diagnostik GmbH,Hessisch Oldendorf,Germany)under the following conditions:initial denaturation for 5 min at94°C followed by 25 cycles of denaturation for 30 s at 94°C,primer annealing for 30 s at 58°C and extension for 35 s at 72°C,and terminated by a final hold at 10°C for all loci(Nakajima et al.,2012).200μL of dye(98%formamide,10 mmol/L EDTA,0.05%pararosaniline)was added to each sample.Reactions were heated up to 72°C for 6 min before loading onto 6%microsatellite gels(Sequagel XR,National Diagnostics,Hull,England).For running a microsatellite gel on the 4300 DNA Analyzer(LI-COR,Biosciences,Germany)manufacturer’s instruction was followed.Base pair lengths obtained from the visual analysis was resolved with previously published allele lengths(Nakajima et al.,2012).
| 图表编号 | XD0040194700 严禁用于非法目的 |
|---|---|
| 绘制时间 | 2019.01.01 |
| 作者 | Xuan-Vy Nguyen、Papenbrock Jutta |
| 绘制单位 | Department of Marine Botany, Institute of Oceanography, Vietnam Academy of Science and Technology、Graduate University of Science and Technology、Institute of Botany, Leibniz University Hannover |
| 更多格式 | 高清、无水印(增值服务) |