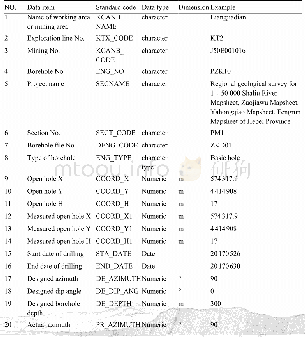
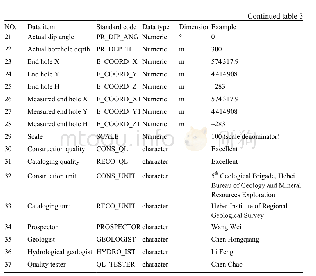
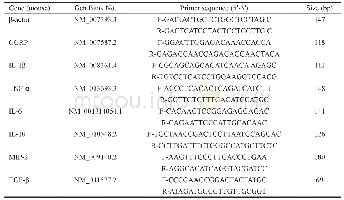
《Table 1.Detailed information for different nucleotides, together with amino acids in the cox1 obtai
 提示:宽带有限、当前游客访问压缩模式
提示:宽带有限、当前游客访问压缩模式
本系列图表出处文件名:随高清版一同展现
《Biased heteroplasmy within the mitogenomic sequences of Gigantometra gigas revealed by sanger and high-throughput methods》
*The heteroplasmic sites which may also lead to heteroplasmic amino acids revealed by Sanger and HTS methods.The amino acids are listed using single-letter amino acid abbreviations.
The presence of heteroplasmy in mitochondrial DNA was reflected in two aspects.On one hand,after iterative check of the chromatograms of all different nucleotides in mitochondrial genes,we found that 74 inconsistent nucleotides are apparently in heteroplasmic sites,of which the second-peak is obvious to eye(Figs 6A–B)and all can be recovered by at least one of the corresponding results of HTS method.Meanwhile,there are other 43 heteroplasmic sites of which the secondpeak is not obvious but can also be recovered by at least one of the corresponding results of HTS method(Figs 6C–D).Moreover,the ratio of such two types of heteroplasmic sites accounts for 94%of all different nucleotides in the coding genes of mitogenome between the two sequencing methods.On the other hand,according to the differently sequenced sites revealed by Sanger and the three assembly sequences of HTS,90%of them are suggested to be heteroplasmy ones at which one of the suboptimal bases is the result reached by Sanger method(Table S6).However,as for the 29 different nucleotides situated in the control region,we cannot detect the discernible second-peak at all.Due to the control region is noncoding gene of mitogenome,we could not rule out the probability of pseudogenes.
| 图表编号 | XD0016210900 严禁用于非法目的 |
|---|---|
| 绘制时间 | 2018.10.01 |
| 作者 | Xiaoya Sun、Yanhui Wang、Pingping Chen、Hesheng Wang、Lixiang Lu、Zhen Ye、Yanzhuo Wu、Teng Li、Wenjun Bu、Qiang Xie |
| 绘制单位 | Institute of Entomology, College of Life Sciences, Nankai University、Department of Ecology and Evolution, College of Life Sciences, Sun Yat-sen University、State Key Laboratory of Biocontrol, Sun Yat-sen University、Netherlands Biodiversity Centre Naturalis |
| 更多格式 | 高清、无水印(增值服务) |