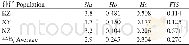《Table 2–Comparison of genetic diversity detected within different mung bean germplasm by SSR marker
 提示:宽带有限、当前游客访问压缩模式
提示:宽带有限、当前游客访问压缩模式
本系列图表出处文件名:随高清版一同展现
《Genetic diversity assessment of a set of introduced mung bean accessions(Vigna radiata L.)》
In contrast to previous studies on mung bean diversity analysis with SSR markers,we observed a higher diversity level,considering the distribution of the germplasm studied(Table 1).In total,the NA ranged from 2 to 6 in most of the diversity analyses of the mung bean[6,7,29,30,32,39,40]as we list in Table 2,and only an average of 6.1 alleles were detected within a core collection of 1481 mung bean accessions[7],indicating low genetic variation in the mung bean compared to the soybean genome[41].We detected an average of 4.2alleles per locus using 184 accessions,a value similar to that detected in 692 accessions from 27 countries[30]and higher than that detected in 157 accessions using EST-SSR markers[29].We also detected a higher average PIC value compared to those in previous reports(Table 2),indicating a greater richness in the distribution of alleles.The GSCs in the present study were in agreement with the calculated diversity values.Most of the GSCs were lower than 0.50 and there were some accession pairs with GSCs of zero,suggesting that they had completely different genetic backgrounds.
| 图表编号 | XD0012319900 严禁用于非法目的 |
|---|---|
| 绘制时间 | 2018.04.01 |
| 作者 | Lixia Wang、Peng Bai、Xingxing Yuan、Honglin Chen、Suhua Wang、Xin Chen、Xuzhen Cheng |
| 绘制单位 | Institute of Crop Sciences, Chinese Academy of Agricultural Sciences、China National Seed Group Co., Ltd.、Institute of Vegetable Crops, Jiangsu Academy of Agricultural Sciences、Institute of Crop Sciences, Chinese Academy of Agricultural Sciences、Institute |
| 更多格式 | 高清、无水印(增值服务) |
查看“Table 2–Comparison of genetic diversity detected within different mung bean germplasm by SSR markers.”的人还看了
-

- Table 2 The codes and sequence of SRAP prim ers used in genetic diversity analysis of Sclerotinia sclerotiorum derived f
-

- Table 1 Comparison of different detection methods and strategies for evaluating PD-L1 expression levels