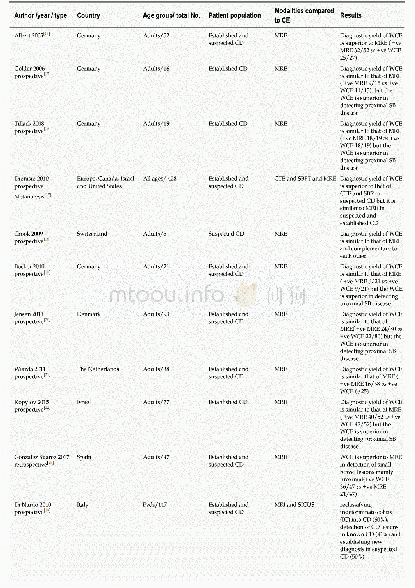
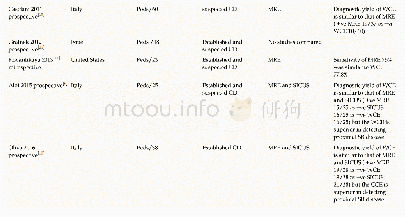
《Table 1–Studies reliably linking structural variation to trait variation in polyploid crops.》
 提示:宽带有限、当前游客访问压缩模式
提示:宽带有限、当前游客访问压缩模式
本系列图表出处文件名:随高清版一同展现
《The role of genomic structural variation in the genetic improvement of polyploid crops》
Copy-number variation can also be understood as an extreme form of sequence variation.Whereas deletions abolish gene copy function,duplications can lead to variation in expression level[13]and thereby affect gene dosage.Duplications thus have a higher risk of affecting traits than point mutations or InDels,and should accordingly be under strong selection pressure.The adaptive value of gene duplicates is illustrated by the observation that polyploid genomes tend to lose redundant gene copies[52],whereas gene copies from adaptive pathways are retained or even duplicated.For example,in the mustard family,genes from the glucosinolate pathway were highly retained after whole-genome duplications,with a retention rate of 95%compared to 45%across all genes[96].Despite the strong phenotypic effects expected from CNV,findings linking genomic variation with phenotypes are scarce.This scarcity is due partly to the comparatively large effort needed to confirm CNVs in plant genomes.CNVs can be detected via microscopy-based approaches,e.g.,fluorescent in situ hybridization(FISH);quantitative PCR(qPCR);probe-hybridization-based assays;SNP array-based methods;or next-generation sequencing.Statistically linking CNVs to phenotypes requires analysis of medium-to-largesized populations,and these must undergo CNV detection as well as phenotyping,which can be expensive in cost and time.Moreover,some of these methods have specific drawbacks.For example,PCR-based approaches are not robust against sequence variation within primer binding sites,SNP arrays cannot easily detect duplications,and NGS approaches depend on the quality of the reference genome.These difficulties partly explain the lack of data.Another reason may be that both scientists and breeders underestimate the extent of structural variation in polyploid crops and focus rather on classical sequence variants.Recent findings of crop genomic structural variation,however,show that the assumption of a stable genome is questionable.This genomic instability is also shown by several pan-genomic approaches.For example,sequencing 10 different accessions of cabbage,a diploid with multiple polyploid events in its lineage(mesohexaploid),revealed that 18.7%of the gene copies were affected by PAV in at least one of the accessions[82].Another pan-genome study in the related recent allotetraploid rapeseed,using 53 diverse accessions including resynthesized lines,found that 38.0%of the genes were affected by PAV[23],similar to the value of 35.7%obtained from 18 hexaploid wheat cultivars[35].These figures illustrate the importance of structural variation in polyploid crops and the pressing need to investigate the influence of structural variants on adaptive and agronomic traits.Although transposable elements are the major source of structural variation in diploids,polyploids present even more possibilities of genomic rearrangement.Homeologous exchanges in meiosis can lead to deletions and duplications in homeologous chromosomes,depending on the degree of sequence similarity and distance from the centromeres[97].A summary of research studies that have confirmed the phenotypic influence of CNVs in polyploid plants is presented in Table 1.
| 图表编号 | XD0029303600 严禁用于非法目的 |
|---|---|
| 绘制时间 | 2019.04.01 |
| 作者 | Sarah-Veronica Schiessl、Elvis Katche、Elizabeth Ihien、Harmeet Singh Chawla、Annaliese S.Mason |
| 绘制单位 | Department of Plant Breeding, Justus Liebig University、Department of Plant Breeding, Justus Liebig University、Department of Plant Breeding, Justus Liebig University、Department of Plant Breeding, Justus Liebig University、Department of Plant Breeding, Justu |
| 更多格式 | 高清、无水印(增值服务) |