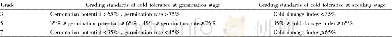《Table 3–Inheritance of abiotic stress tolerance in chickpea and pigeonpea.》
 提示:宽带有限、当前游客访问压缩模式
提示:宽带有限、当前游客访问压缩模式
本系列图表出处文件名:随高清版一同展现
《Integrated physiological and molecular approaches to improvement of abiotic stress tolerance in two pulse crops of the semi-arid tropics》
Polymerase chain reaction(PCR)-based simple sequence repeats(SSR)and single-nucleotide polymorphism(SNP)markers have frequently been used for their ease in genotyping of large segregating populations in a cost-effective manner with high reproducibility.In chickpea,>44,000 ESTs and 2000 genomic SSR markers have been reported[108].Gaur et al.[109]reported nearly 800 SSR markers;however,the frequency of polymorphic markers was quite low(30%)as compared to that in other genera[110,111].International Crops Research Institute for the Semi-Arid Tropics(ICRISAT),in association with University of Frankfurt(Germany)and University of California(Davis,USA),has developed 311 SSR markers from SSR-enriched libraries and 1344 SSR markers from BAC-end sequence in chickpea using mining approaches[111].A 738-Mbp draft whole genome shotgun sequence of a kabuli chickpea variety‘CDC Frontier’comprising 28,269 genes,several unigenes,SSRs,and SNPs(Table 4)has been reported[112].A comprehensive genetic map of chickpea was developed by several researchers using RIL populations of ICC 4958×PI 489777[109–111].Although saturated linkage maps are available,mapping and tagging of biotic and abiotic stress-tolerance loci with tightly linked markers are still lacking.There are reports of many EST-based sequences that are tissue-and developmental stage-specific[113].In addition,transcriptomic and proteomic studies have resulted in the identification of various stress-responsive ESTs and genes[114].Garg et al.[115]reported dynamic transcriptional responses of chickpea tissues to various abiotic stresses and identified 11,640 chickpea transcripts.Differential expression analysis of these transcripts revealed response to at least one of three(limited moisture,salinity and chilling)abiotic stresses.The details of other genomic resources[116–121]are presented in Table 4.
| 图表编号 | XD0012312900 严禁用于非法目的 |
|---|---|
| 绘制时间 | 2018.04.01 |
| 作者 | Arbind K.Choudhary、Rafat Sultana、M.Isabel Vales、Kul Bhushan Saxena、Ravi Ranjan Kumar、Pasala Ratnakumar |
| 绘制单位 | ICAR Research Complex for Eastern Region、Bihar Agricultural University、Texas A&M University, College Station、International Crops Research Institute for the Semi-Arid Tropics、Bihar Agricultural University、National Institute of Abiotic Stress Management |
| 更多格式 | 高清、无水印(增值服务) |