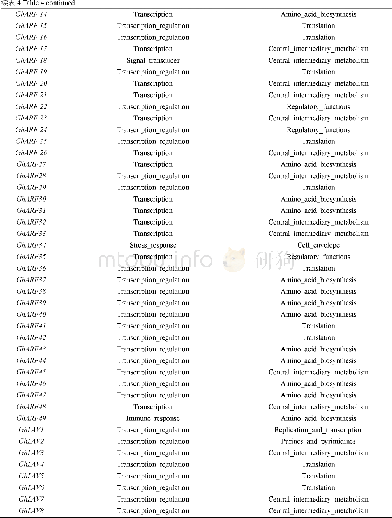
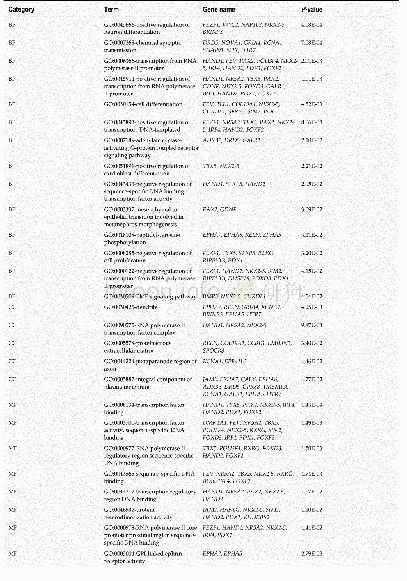
《Table 1 Gene ontology enrichment analysis of methylation-regulated differentially expressed genes a
 提示:宽带有限、当前游客访问压缩模式
提示:宽带有限、当前游客访问压缩模式
本系列图表出处文件名:随高清版一同展现
《Identification of differentially expressed genes regulated by methylation in colon cancer based on bioinformatics analysis》
BP:Biological process;CC:Cellular component;MF:Molecular function.
We further performed functional enrichment analysis to clarify the role of methylation in colon cancer.In biological process enrichment of GO,as many as 12terms were related to the regulation of transcription.Therefore,we speculated that transcription factors regulated by methylation can further regulate the transcription of cancer-related genes and thus affect the occurrence and development of cancer.Aberrant cell proliferation is regarded as the distinguishing feature of cancer cells as opposed to normal cells.The term“negative regulation of cell proliferation”was enriched,meaning that multiple cancer suppressor genes were down-regulated by promoter hypermethylation.As shown in Table 1,MeDEGs including FEZ family zinc finger protein 1(FEZF1),T-box transcription factor 5(TBX5),secreted frizzled-related protein 5(SFRP5),RAS-like estrogen regulated growth inhibitor(RERG),ripply transcriptional repressor 3(RIPPLY3),and pancreatic and duodenal homeobox 1(PDX1)were involved.Among them,TBX5[23],SFRP5[24,25],RERG[26-28],and PDX1[29]have been reported to be regulated by methylation in colon cancer,while FEZF1 and RIPPLY3 have not been reported.Bone morphogenetic proteins(BMPs),which form part of the transforming growth factorβsignaling pathway,can promote colon cancer cell migration and invasion[30,31].Moreover,methylation of BMP3[32,33]and NK2homeobox 5[34]promoters has been revealed as an independent biomarker for colon cancer.However,apart from colon cancer,there have been no reports on the role of CHRDL1 promoter region methylation in tumors[35].KEGG pathway analysis has further clarified the function of MeDEGs.The“cAMP signaling pathway”has been indicated as one of the most critical mechanisms in regulating colon cancer cell apoptosis[36-38].The term“transcriptional misregulation in cancer”is consistent with the GO enrichment results.Babykutty et al[39]indicated that“cGMP-PKG signaling pathway”could promote colon cancer cell migration and invasion by upregulating matrix metalloprotein 2/9.
| 图表编号 | XD0087246900 严禁用于非法目的 |
|---|---|
| 绘制时间 | 2019.07.14 |
| 作者 | Yu Liang、Cheng Zhang、Dong-Qiu Dai |
| 绘制单位 | Department of Gastrointestinal Surgery, the Fourth Affiliated Hospital of China Medical University、Department of Gastrointestinal Surgery, the Fourth Affiliated Hospital of China Medical University、Department of Gastrointestinal Surgery, the Fourth Affili |
| 更多格式 | 高清、无水印(增值服务) |
查看“Table 1 Gene ontology enrichment analysis of methylation-regulated differentially expressed genes associated with colon”的人还看了
-

- Table 2 Kyoto encyclopedia of genes and genomes pathway analysis of methylation-regulated differentially expressed genes