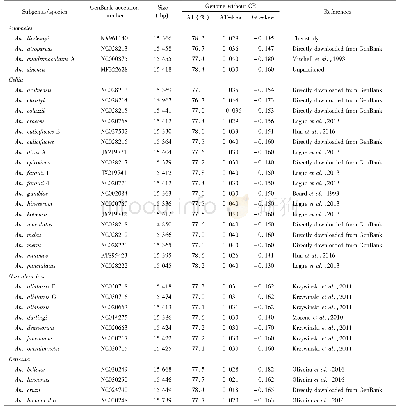
《Table 1 Accession numbers and information of mtgenomes used in the present phylogenetic analysis》
 提示:宽带有限、当前游客访问压缩模式
提示:宽带有限、当前游客访问压缩模式
本系列图表出处文件名:随高清版一同展现
《林氏按蚊线粒体全基因组序列的测定及基于线粒体基因组的按蚊属系统发育分析(英文)》
The phylogenetic relationships of 33 mtgenome sequences of Anopheles were analyzed using both maximum likelihood(ML)and Bayesian inference(BI)methods with the mtgenome sequence of Culex pipiens pallens(KT851543)as the outgroup(Table1).Among all of the 33 mtgenome sequences,the mtgenome of An.lindesayi was newly sequenced in this study,that of An.sinensis has earlier been reported(Hao et al.,2017)but not yet available in the GenBank,and all the other 31 mtgenome sequences were downloaded from the GenBank.The nucleotide and amino acid sequences of 13 proteincoding genes were separately aligned using MAFFT(Kazutaka and Standley,2013).After removing the poorly aligned and divergent regions using Gblocks(Talavera and Castresana,2007),the individual alignments were then concatenated following their orders in the mitogenome.The best-fit substitution model for nucleotide datasets and AA datasets were selected by Modeltest using the Akaike Information Criterion(AIC)(Posada and Crandall,1998) and ProtTest,and the GTR+I+G and WAG+G+I+F were determined as the best-fit models for nucleotide and AA sequences,respectively.RAx ML(Stamatakis,2014)was used in the ML analysis under the best-fit model detected,and the bootstrap support values were calculated based on 1 000 replicates.MrBayes(Ronquist and Huelsenbeck,2003)was used in the BI analysis with the two independent runs and four chains(three heat and one cold)were performed simultaneously for 1 000 000 generations.The runs were stopped till the average deviation of split frequencies fall below 0.01,and the phylogenetic trees being sampled every 100 generations with 25%burn-in rate.The topology of the best-scoring trees were visualized and edited in MEGA 5.Bootstrap support values larger than 50%,and the names of four subgenera and eight Series in the genus Anopheles were marked on the corresponding branches of the constructed trees.
| 图表编号 | XD0036515400 严禁用于非法目的 |
|---|---|
| 绘制时间 | 2019.01.20 |
| 作者 | 毛启萌、李廷景、付文博、闫振天、陈斌 |
| 绘制单位 | 重庆师范大学昆虫与分子生物学研究所重庆市媒介昆虫重点实验室、重庆师范大学昆虫与分子生物学研究所重庆市媒介昆虫重点实验室、重庆师范大学昆虫与分子生物学研究所重庆市媒介昆虫重点实验室、重庆师范大学昆虫与分子生物学研究所重庆市媒介昆虫重点实验室、重庆师范大学昆虫与分子生物学研究所重庆市媒介昆虫重点实验室 |
| 更多格式 | 高清、无水印(增值服务) |





