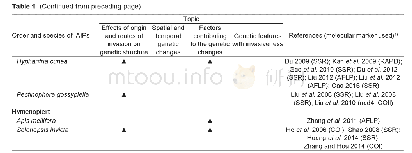
《Table 1–cis-regulatory elements involved in endosperm expression.》
 提示:宽带有限、当前游客访问压缩模式
提示:宽带有限、当前游客访问压缩模式
本系列图表出处文件名:随高清版一同展现
《"Imprinting, methylation, and expression characterization of the maize ETHYLENE-INSENSITIVE 2-like gene"》
The sequences of the gene and its predicted promoter region were downloaded from http://www.maizesequence.org/index.html.The gene was amplified from B73 using KOD FX DNA polymerase(Toyobo,Osaka,Japan).Rapid amplification of cDNA ends was used to determine the transcriptional start site(TSS)and 3′untranslated region(UTR)of EIN2-like.Cis-regulatory elements were predicted by Plant CARE(http://bioinformatics.psb.ugent.be/webtools/plantcare/html/)[32]and Cp G islands were predicted by the EMBOSS Cpgplot program(http://www.ebi.ac.uk/Tools/seqstats/emboss_cpgplot/).Multiple sequence alignments were performed using Clustal X1.83,as implemented by MEGA 5.0,on full-length proteins[33].Phylogenetic trees were constructed by neighbor joining with the JTT model and Complete Deletion[33].Reliability values at each branch point represent bootstrap cores(1000 replicates).Conserved motifs of the EIN2 protein sequences were identified by MEME(http://meme-suite.org/tools/meme).The sequences of all the primers used in the study are listed in Table S1.
| 图表编号 | XD0029301300 严禁用于非法目的 |
|---|---|
| 绘制时间 | 2019.02.01 |
| 作者 | Xiupeng Mei、Ping Li、Lu Wang、Chaoxian Liu、Lian Zhou、Chunyan Li、Yilin Cai |
| 绘制单位 | Maize Research Institute, Key Laboratory of Biotechnology and Crop Quality Improvement, Ministry of Agriculture, Southwest University、Maize Research Institute, Key Laboratory of Biotechnology and Crop Quality Improvement, Ministry of Agriculture, Southwes |
| 更多格式 | 高清、无水印(增值服务) |
查看“Table 1–cis-regulatory elements involved in endosperm expression.”的人还看了
-
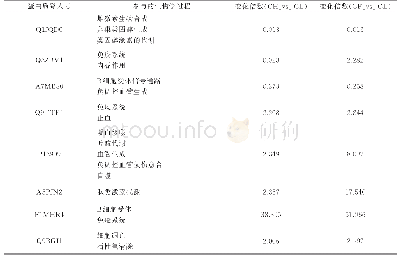
- 表3 可能与雌性水牛发情周期相关的重要差异表达蛋白质Table 3 The important proteins that may be involved in the estrus cycle of female buffalo
-
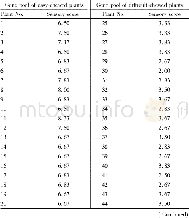
- Table 2Sensory evaluation of Chinese cabbage plants involved in the verification of RAPD primer OPA-06
-

- Table 2Sensory evaluation of Chinese cabbage plants involved in the verification of RAPD primer OPA-06